Up to FastTree
FastTree can constrain its search for the best tree to try and satisfy constraints that you give it. Each constraint takes the form of a split. For example, if you constrain A and B versus C and D, then FastTree will avoid topologies such as ( (A,C), B, (D,E) ) that do not contain that split. (The placement of E is ignored, as it was not specified in the constraint.) You can specify any number of constraints, although too many constraints will make FastTree much slower: roughly speaking, each constraint costs as much as adding a few nucleotide positions to the multiple sequence alignment. Each constraint can contain any number of leaves, but a split that has less than two taxa on a side is not meaningful because every tree satisfies that split.
FastTree expects you to give it constraints in the form of a constraint alignment, rather than that of a tree. Each column in the constraint alignment contains a mix of "0", "1", and "-" characters. The taxa with 0 and 1 values are required to be on opposing sides of a split. The constraint alignment can be in either phylip interleaved or fasta format. For example, if you want to constrain the splits AB:CD and AC:DE, then your constraint alignment would be
5 2 A 00 B 0- C 10 D 11 E -1You can use this constraint file together with a multiple sequence alignment, e.g.
FastTree -constraints constraint_alignment < multiple_sequence_alignment > treeNot all of the sequences that are in the MSA need to be present in the constraints file.
One common way to get constraints for constructing a large tree with FastTree is from a smaller tree that does not contain all of the taxa of interest but is of high quality. You can use the TreeToConstraints.pl perl script to convert a tree to a constraint alignment, e.g.,
perl TreeToConstraints.pl < small_tree > constraint_alignmentThis script requires both perl and BioPerl to be installed.
Constraints can be violated for many different reasons:
Unless you run FastTree with the -quiet option, it will report how successfully it has met the constraints like this:
Bad splits by min. evo.: 102 of 1223 violating constraints: 0 both bad: 0According to this report, none of the constraints were violated. "Bad" splits are locations in the tree where a nearest-neighbor interchange would reduce the length of the tree -- if you run FastTree with constraints, then there will be more of these than normal. (Note that the maximum likelihood phase of FastTree may correctly introduce splits that lengthen the tree but increase the likelihood.)
The -constraintWeight option controls how strongly FastTree prefers to satisfy a constraint rather than reducing the length of the tree. By default, the constraint weight is 10.0. In other words, a join that causes one additional constraint to be violated is treated as if it lengthened the tree by 10.0 more than it actually does. During the maximum likelihood phase, it treats a violation as equivalent to a decrease in the log likelihood of 10.0. These may sound like a lot -- trees don't normally have branch lengths of 10! But if a constraint can be violated partially -- that is, most of the sequences split correctly, and just one is out of place. In that case, FastTree will only assign a fractional penalty. Thus, without a high constraint-weight, FastTree may not satisfy all of the constraints that it could.
FastTree tries to satisfy the constraints at every step of neighbor-joining, and it tries to improve the agreement of the tree with the constraints at every nearest-neighbor interchange or subtree-prune-regraft move. In other words, it uses a greedy algorithm to find a good topology, and then it tries local changes to improve the topology.
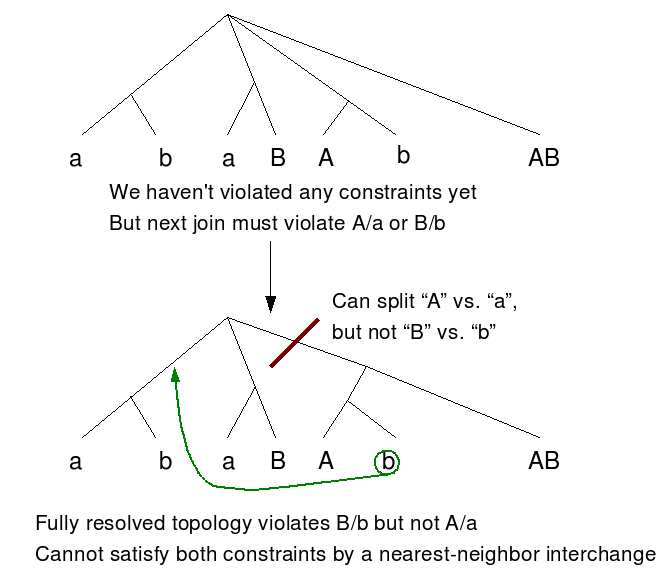
This example shows that this approach may fail to converge on a topology that satisfies the constraints, even though such a topology exists. The taxa are labeled with their constraints (A/a and B/b are the two constrained splits). At the top of the figure, the tree is partially resolved because neighbor joining has not completed yet. Neither constraint has been violated in the partially resolved tree, but either of the two possible fully resolved topologies will violate a constraint. One of the problematic topologies is shown below.